This week we released version 2.0 of PathVisio. There has been over a year of active development since the last major release, and a ton of new features.
What is PathVisio? PathVisio is a tool for biological pathways. Stay organized! Use PathVisio as a simple notebook to collect all the various bits of information related to a biological research subject. Create images suitable for presentation or publication. Draw pathways, export them to many image formats, annotate them with links to online biological databases such as Ensembl or Entrez gene, and add comments and literature references from pubmed.
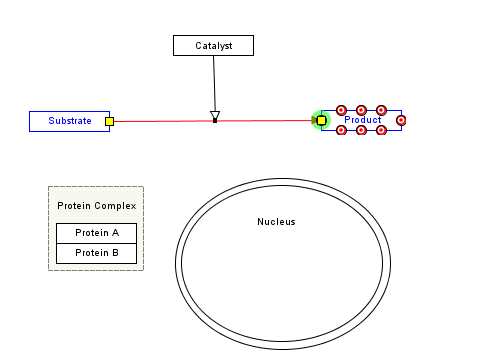
With PathVisio you draw pathways just like you would in powerpoint:
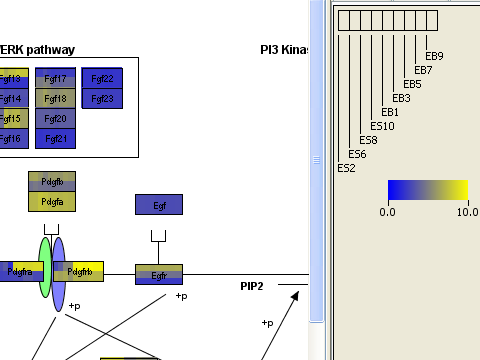
What is new? New in PathVisio 2.0 is the ability to import experimental datasets and visualize them on top of pathways. Explore large datasets in a way that is more interesting and understandable than just a huge spreadsheet. Import microarray, proteomics or metabolomics data. Microarray reporters will be automatically linked to genes and protein identifiers in pathways. You can customize the visualization, using gradients, boolean color rules, or colored icons.
Here is an example of visualized microarray data:
Perform over-representation analysis to find the pathway that was most affected by experimental conditions. This is great for hypothesis-generating experiment types, where you really don’t know anything in advance about your experiment.
Download pathway sets or share pathways on WikiPathways, a wiki where any researchers can contribute pathway knowledge. PathVisio is fully compatible.
Check our visual tour if you want to know more. Click here to Download PathVisio
For Developers: PathVisio has a plugin interface that lets users customize it to new analysis types, new visualization methods and new pathway formats. PathVisio is fully open source, and we’re always looking for Java developers who are interested in contributing, either to new plugins or to the core of the program. Contact us on our mailinglist.
Tags: announcement, pathvisio